Cell Motility - The Crawling Mechanism Of Actin Assembly
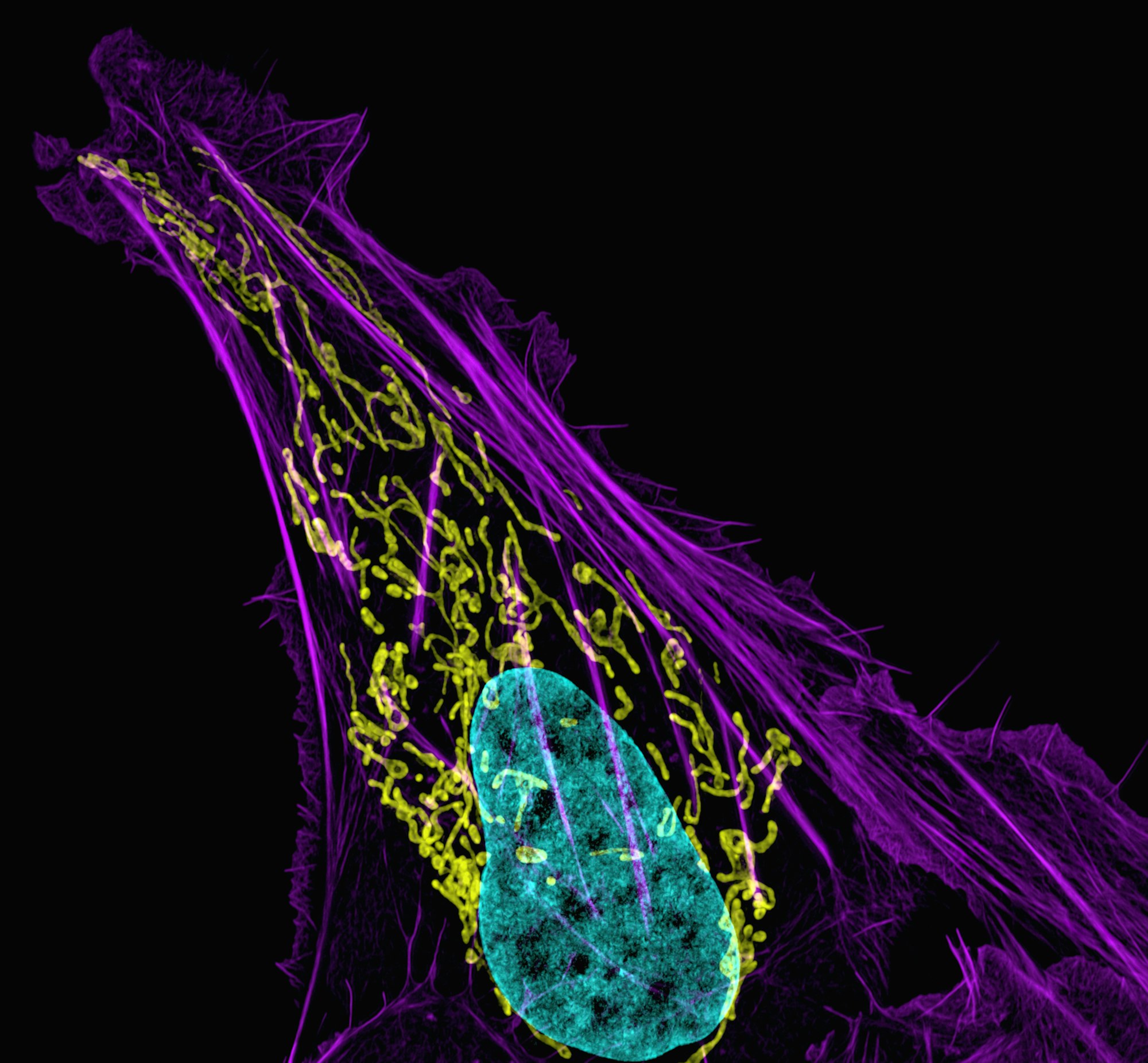
Cells, when going about their everyday business, do not think. Like a great symphony orchestra played by a million synchronised musicians, cells might appear to carry an intelligent consciousness to them - though of course they do not. They are mindless machines that function based on the incredible chemistry and physical properties that make them, only able to react to their changing environment as a result of evolution (with millions of years of natural selection producing cells that are able to survive and reproduce most effectively). The typical cell, for example, has a multitude of assigned signalling proteins and enzymes to regulate resource consumption and waste excretion. Each of these work as certain chemical thresholds are reached (e.g., when there are sufficient autoinducers to trigger quorum sensing in bacterial populations), which in turn alter protein conformations to cause protein interactions or control gene expression in the cell.
My professor once told me that cells use just as much energy to hold back reactions from occurring as they do in activating them. Whether this is strictly true or not, I am not sure - but the message is clear. Given the constant flux of their surroundings, it is crucial for cells to be capable of formulating appropriate responses to them as quickly as possible. To do this, they have evolved mechanisms that are so energetically favourable and 'ready to go' that they can be triggered almost instantaneously in case they need them, and it is such processes that further regulate how cells move. They are quite complex, however. As such, while the movement systems in both eukaryotic and prokaryotic cells are endlessly fascinating to explore, we will stick with the former for this post. The point of study: a phagocytic macrophage chasing after its prey.
The Crawl
To compete with the vivacious swimming of a fleeing bacterium - which uses the awesome power of rotating flagellar motors to travel - the bulkier macrophage must have a far more widely-encompassing mechanism to transport its body across a tissue matrix (i.e., the material between an organism's cells). Fortunately, eukaryotic cells have developed a trick for this: the cytoskeleton. Different from the one in bacteria, the eukaryotic cytoskeleton is constituted primarily of actin filaments, microtubules and intermediate filaments. These are made of different kinds of proteins, with some only occurring in specific locations on the organism (such as collagen intermediate filaments in animal cells), but they work together to provide both structure and support in animal cells. They also allow the transport of resources inside cells, further inducing the rapid locomotion that we observe in crawling macrophages.
Actin filaments are particularly relevant in this case. Having first been found alongside myosin in 1942 as the main proteins involved in muscle contraction by biochemists Brúno Straub and Ilona Banga, actin contributes to a vast array of cellular processes (such as endocytosis) and is a model molecule for molecular biologists. Quite simply, this is because of their propensity for polymerisation, which is where multiple subunits repeatedly stack onto each other to form a longer mass (i.e., a polymer). Actin filaments are double helix structures, and they are polymers of - you guessed it! - actin subunits. How these actually polymerise is more interesting, but you can essentially imagine it as a ladder being built by workers piecemeal together at one end to elongate, with the rear end slowly coming undone. The rate at which this occurs changes over time, and eventually even backtracks when the filament needs to disassemble. Either way, many auxiliary proteins work to regulate actin polymerisation - starting with the sequestering protein, thymosin.

At the conditions present in a human body cell, the concentration of globular actin monomers (also known as G-actin) must be approximately 0.2μM for it to polymerise. Cells, however, usually have constant concentrations of up to 70μM - far beyond what is needed for the reaction to occur. Why do we not see filamentous actin (F-actin) all the time, then? See, most of the time, cells require G-actin to remain readily available in case it is required to form filaments at different parts of the cell. Monomers are therefore 'stored' by binding to thymosin, which has a great affinity to G-actin and keeps it from polymerising in the cell. Another crucial aspect of actin is its ability to bind to and hydrolyse adenosine triphosphate (ATP), a molecule that can be hydrolysed to be turned into adenosine triphosphate (ADP). It is in this state - wherein actin is bound to ATP - that thymosin keeps it.
When the macrophage starts chasing the bacterium (say, due to a chemical signal that it sensed), a protein known as profilin binds to free G-actin in the cytosol and thereby brings it to a nucleator. There are a number of protein complexes that can act as a nucleators in different cells (such as formins or ARPs), but all of them have essentially the same purpose: to catalyse the polymerisation of actin. Once profilin has led the monomer to a nucleator, it can then quickly form a base for other G-actin monomers to stack on top of, at which point thymosin dissociates with G-actin (as its concentration decreases) and causes elongation to occur spontaneously and faster than you could say 'wow!' In the right conditions, it even looks like mini-fireworks all moving concurrently and irrepresably in the cell, and I think it is absolutely spectacular.
Of course, we do not want actin to continue elongating forever. (It would be quite a lot of trouble otherwise!) The cell does not want this, either, so after actin's elongation phase comes a steady phase. Here, the actin filaments have grown long enough that they stay at a constant length. Instead of simply stopping, however, the filaments are simply in a dynamic equilibrium as a result of actin's role as an ATPase. Over time, F-actin causes ATP bound to it becomes hydrolysed into ADP, which in turn makes it more likely to dissociate from the filament. Since actin polymerises more easily at only one end of the filament (barbed end), F-actin bound to ADP accumulates at the rear (pointed end). After a certain point, this means that actin filaments keep on growing as much at the barbed end as they are losing subunits on the pointed end - and so we give it the name treadmilling. Actin treadmilling results in filaments of constant length moving steadily in one direction.
As the actin mesh forms and shoots forward, it ultimately hits and pushes the cell membrane in the direction of travel. Like dough, the membrane stretches to form outward lamillipodia, reaching for a pathogen in the case of the macrophage. It then creates cell-matrix junctions to adhere to the surroundings (preventing it from rebounding like a rubber band) and breaks down the junctions at its trailing end. The motion repeats itself without end - or at least until the cell has moved to a more favourable state, or its energy in the form of ATP has run out. Consequently, the overall motion produced looks very similar to a crawl, which admittedly does not sound pace-breakingly fast. Albeit, it suits the cell just fine; in the end, the macrophage ends up devouring its fleeing prey only seconds after being spotted.
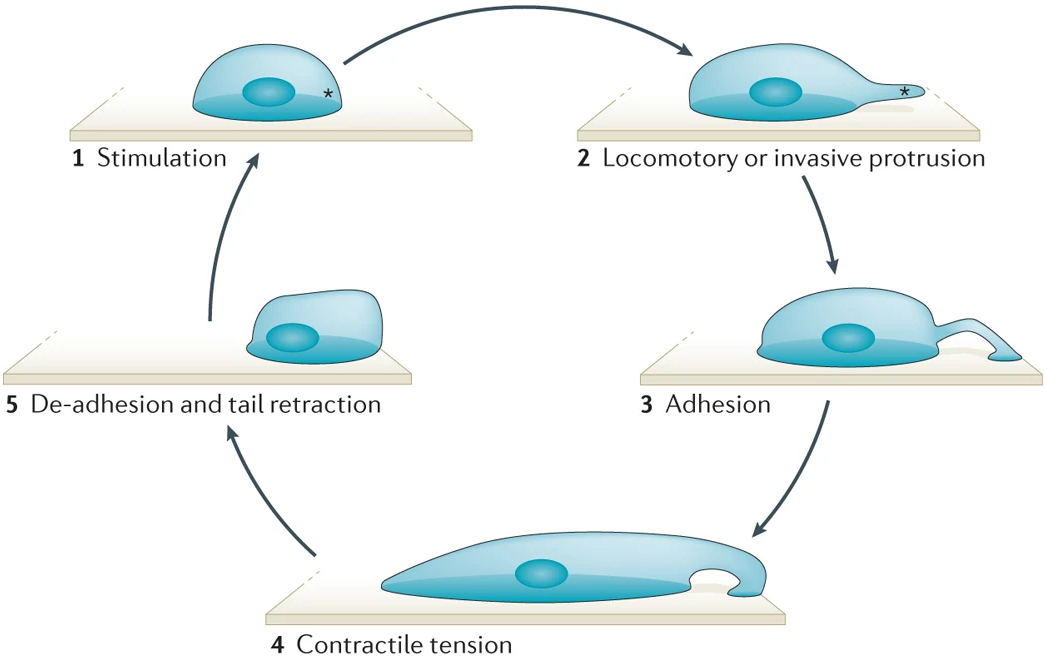
After the cell is done moving, it needs to revert its cytoskeleton to how it was before. Fortunately, the modular semblance of actin filaments allows rapid and efficient deconstruction whenever necessary, allowing the cell to either store the actin monomenrs with thymosin again or reuse them for another purpose. There are a couple other players in the depolymerisation of actin filaments, however, as we shall explore below.
Slowing Down To A Stop
Eukaryotic cells have a number of mechanisms to shift their speed and direction in only moments. Given their gigantic size (relative to biological molecules), this mainly involves transferring their internal cytoskeleton around to 'push' from a different side, assisted by the spherical symmetry of the cell. Not only is this far quicker than having to turn the entire figure around, it also conserves significantly more energy. With this in mind, we can see why using actin filaments to move is so convenient - they are easily built, strong and collapsible in a pinch.
One method by which actin filaments can depolymerise is by the hydrolysis of bound ATP (as we saw above). Lowering the affinity for F-actin to remain in the filament, this is integral in treadmilling and acts as the basic framework for other such mechanisms. When activated, cofilin proteins are then prone to bind to and sever filaments into smaller pieces, either causing them to depolymerise more quickly or producing a greater number of elongating strands to have a more widespread effect on the cell. Cofilin activity is regulated by a range of accessory proteins, triggered only when a cell requires them to work.
I should note that F-actin dissociation does not only occur at the pointed end of the filament. In fact, contrary to intuition, it happens even more commonly at its barbed end! Nonetheless, since the rate of association is so much higher here, we typically don't think too much of it. (As the average science teacher would say, just ignore it for now.) Moreover, CAP proteins that associate at the barbed end of a filament usually stabilise them to keep them from desintigrating spontaneously, hence increasing polymerisation there. In the same way, Mical proteins are known to be involved in the dynamic disassembly of actin filaments. Apparently, Mical behaves as an oxidoreductase and is able to oxidise F-actin at the barbed end of a chain, permanently disabling their ability to bind to further monomers and ultimately breaking it down. Recent research has further established that Mical-oxidised G-actin still binds to profilin, thereby inhibiting profilin from transporting other available G-actin and indirectly blocking actin polymerisation as a whole.
Now, there are quite a number of other players in build-a-block actin (dis)assembly, but the field gets pretty deep and pretty convoluted fast. Biomechanics research is particularly interested in this - for example when trying to understand the reasons behind the canine metabolism and diseases - and it often integrates a significant amount of more exact calculations. Albeit, knowing that cells move around this way is more than a good starting point, particularly since it serves as good model for future nanotech. It also acknowledges the complexity of cell quite nicely, I believe!
References
- Bugyi, B., Kellermayer, M. (2020). The discovery of actin: “to see what everyone else has seen, and to think what nobody has thought”. Journal of Muscle Research and Cell Motility 41:3-9. Retrieved from https://doi.org/10.1007/s10974-019-09515-z
- Lasso, D. M., et al (2020). Much More Than a Scaffold: Cytoskeletal Proteins in Neurological Disorders. Cells 9(2):358. Retrieved from http://dx.doi.org/10.3390/cells9020358
- Boyle, S. & Kopecki, Z. (2019). Mechanical Force and Actin Dynamics during Cutaneous Squamous Cell Carcinoma (cSCC) Progression: Opportunities for Novel Treatment Modalities. Squamous Cell Carcinoma - Hallmark and Treatment Modalities. Retrieved from http://dx.doi.org/10.5772/intechopen.86041
- Bugyi, B. & Carlier, M. (2010). Control of Actin Filament Treadmilling in Cell Motility. Annual Review of Biophysics. Retrieved from https://doi.org/10.1146/annurev-biophys-051309-103849
- Bravo-Cordero, J., et al (2013). Functions of cofilin in cell locomotion and invasion. Nature Reviews Molecular Cell Biology 14:405–415. Retrieved from https://doi.org/10.1038/nrm3609
- Grintsevich, E. E., et al (2021). Profilin and Mical combine to impair F-actin assembly and promote disassembly and remodeling. Nature Communincations 12:5542. Retrieved from https://doi.org/10.1038/s41467-021-25781-3
